Our lab investigates genes and networks regulating muscle identity, muscle size, and muscle organelle positioning, particularly myonuclear movement and placement.
We use Drosophila and, more recently, human progenitor stem cell (hPSCs)- derived muscle cells as our model systems.
In the Drosophila embryo, a repeated pattern of 30 distinct muscle fibers is present in each abdominal hemisegment. Despite similarities, such as shared expression of contractile proteins, each muscle fiber can be distinguished by properties like its size, shape, orientation, number of nuclei, innervation, and tendon attachment site (Figure 1). We identify key processes in muscle development and differentiation in Drosophila and then test these in human muscle cells.
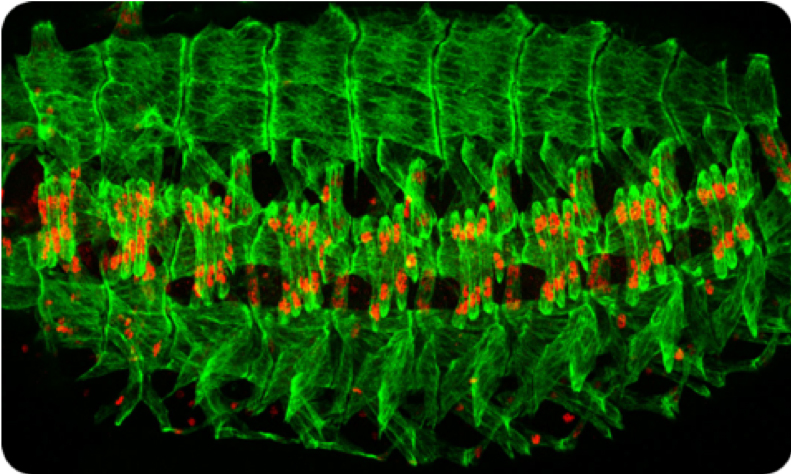
Figure 1: The body wall musculature of the Drosophila embryo. This image shows a late stage Drosophila embryo in which the individual muscle cells (green) are visualized. There is a repeated pattern of muscle cells, with 30 per hemisegment. Red stain reveals the individual nuclei present in a subset of muscle cells.
Selected reviews from the lab:
Dobi KC, Schulman VK, Baylies MK. Specification of the somatic musculature in Drosophila. Wiley Interdiscip Rev Dev Biol. 2015 Jul-Aug;4(4):357-75. doi: 10.1002/wdev.182. Epub 2015 Feb 27. PMID:25728002
Schulman VK, Dobi KC, Baylies MK. Morphogenesis of the somatic musculature in Drosophila melanogaster. Wiley Interdiscip Rev Dev Biol. 2015 Jul-Aug;4(4):313-34. doi: 10.1002/wdev.180. Epub 2015 Mar 11. PMID:25758712
Muscle identity: transcription and chromatin regulation
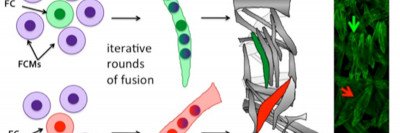
Muscle fibers arise by the iterative fusion of two types of myoblasts, called founder cells (FCs) and fusion competent myoblasts (FCMs), to form a syncytium. FCs are thought to contain all the information needed to make a muscle of a particular size and shape. This information is encoded by DNA-binding transcription factors, such as Krüppel, Slouch, and Apterous, expressed in incompletely overlapping subsets of FCs, known as the FC identity genes.
Using a novel molecular screen, we have identified new transcription factors, such as Alhambra and Charlatan, and, for the first time, chromatin regulators such as Sin3a and Kdm2 that are enriched in different populations of FCs (Figure 2). We have found that the chromatin regulators buffer the function of the transcriptional regulators, assuring specific muscle identities. Current investigation is directed at finding how morphological information is translated from the identity regulators to the cellular processes that control muscle size and shape.
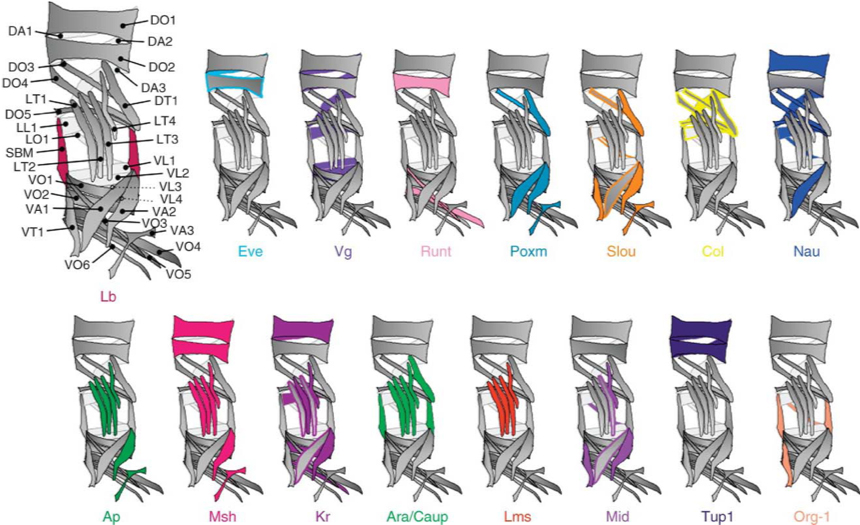
Figure 2: Muscle transcriptional genes control the muscle identity (size, shape, orientation, innervation) through differential gene regulation. These identity factors are expressed in different subsets of muscle cells.
Selected publications:
Dobi, KC, Halfon MS, Baylies MK. 2014. Whole-genome analysis of muscle founder cells implicates the chromatin regulator sin3a in muscle identity. Cell Reports. 2014 Jul 30. pii: S2211-1247(14)00572-5. PMID:25088419. PMCID: PMC4207094 doi:10.1016/j.celrep.2014.07.005.
Kumar RP, Dobi KC, Baylies MK, Abmayr SM. 2015. Muscle cell fate choice requires the T-box transcription factor midline in Drosophila. Genetics. 199(3):777-91. doi: 10.1534/genetics.115.174300. Epub 2015 Jan 21. PMID:25614583. PMCID: PMC4349071
Muscle cell size
Cell-cell fusion: membrane dynamics and actin regulation
As in humans, muscle cells in Drosophila form by the fusion of myoblasts. Thirty FCs seed the abdominal muscle pattern and recruit neighboring FCMs to form multinucleate muscle fibers. Certain muscles stereotypically contain as few as three nuclei (indicative of two fusion events), whereas others consistently incorporate up to 25 nuclei (24 fusion events).
Using genetic screens and time-lapse videography, we have identified an actin structure — the F-actin focus — that marks the fusion site in Drosophila. The F-actin focus forms the backbone of an invasive podosome-like structure that is required for fusion to progress. Using different genetic screens, we have shown that this structure can be modulated by several additional actin regulators such as the Formin, Diaphanous, as well as by PI(4,5)P2 signaling (Video 1).
Current experiments are aimed at identifying additional components of the fusion process as well as the mechanism by which muscle cells “count” and determine the characteristic number of fusion events for a particular muscle.
Selected publications:
Bothe, I., Deng, S., Baylies MK. 2014. PI(4,5)P2 regulates myoblast fusion through Arp2/3 regulator localization at the fusion site. Development. 141(11):2289-301. PMID: 24821989. PMCID: PMC4034421
Deng S, Bothe I, Baylies MK. 2015. The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation. PLoS Genet. 2015 Aug 21;11(8):e1005381. doi: 10.1371/journal.pgen.1005381. eCollection 2015 Aug. PMID:26295716
Nuclear Scaling and Nuclear Coordination:
Additionally, we have begun to tackle the question of muscle cell size through quantification and manipulation of different nuclear parameters within a multinucleated muscle cell. For these experiments, we have focused on the musculature of the Drosophila larvae (Figure 3, Video 2). Muscles in the larvae enable its movement and grow 20-40 fold over the course of a 5 day period. This rapid growth is not fueled by the addition of more nuclei as is done in human muscle cells, but by endoreplication (increasing DNA content in each nucleus). Our data indicate that there is coordination among the individual nuclei to generate a specific muscle cell size (global nuclear scaling). However, each nucleus within the muscle cells adjust their size and synthetic activity in correlation with positional or spatial cues (local scaling). While individual myonuclei can exhibit compensatory potential, we find that even minor changes in global scaling correlates with reduced muscle function.
Current projects are focused on determining how nuclei cooperate and compensate as well as establishing the local cues that influence the behavior of individual nuclei.
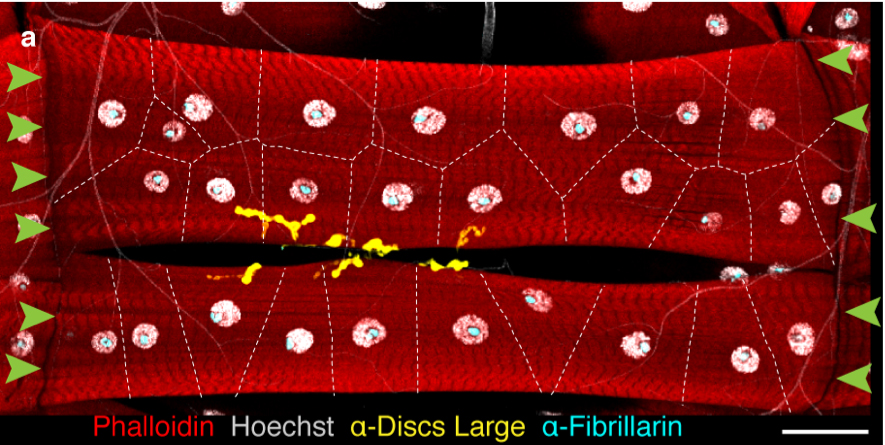
Figure 3: Drosophila Larval muscle cells, VL3 and VL4. High magnification of two individual muscle cells. White, DNA in the nuclei; Cyan, nucleolus; yellow, the NMJ; Green arrows, MTJ. Dotted lines indicate Voronoi domains. We use these muscles to identify different aspects of local and global nuclear scaling.
Selected publications:
Windner SE, Manhart A, Brown A, Mogilner A, Baylies MK. 2019. Nuclear Scaling Is Coordinated among Individual Nuclei in Multinucleated Muscle Fibers. Dev Cell. 49(1):48-62.e3. doi: 10.1016/j.devcel.2019.02.020. Epub 2019 Mar 21. PMID:30905770. Cover image and preview!
Muscle subcellular organization:
Nuclear positioning and cytoskeletal organization
Highlighting the importance of proper intracellular organization, many muscle diseases are characterized by mispositioned myonuclei. We have found that proper positioning of myonuclei is dependent upon microtubule organization, microtubule-associated proteins such as Ensconsin and Ninein, and the microtubule motor proteins Kinesin-1 and cytoplasmic Dynein and its associated proteins, such as Syd, Clip-190, and Glued. Mutations in these proteins affect myonuclear position and muscle function (Figure 4). We have identified at least two distinct mechanisms by which these proteins move and position myonuclei. These motors exert forces both directly on the nuclear surface and from the cell cortex via microtubules.
Current projects investigate additional components required for myonuclear movement, the contributions of other tissues to nuclear placement, and the physiological changes that occur in muscle with displaced nuclei. In addition, we are using mathematical modeling, in collaboration with Dr. Alex Mogilner, to investigate the mechanisms of positioning in all stages of Drosophila muscle development.

Figure 4: Overexpression of Bsg25D (Ninein) affects microtubule organization and myonuclear positioning in the Drosophila larval muscles. Blue, Nuclei; white Microtubules. Top panels: a portion of the VL4 muscle in the genotypes listed. Bottom panels: high magnification of representative nuclei from the genotypes listed. Red arrows indicate microtubules missing from the nuclear envelope.
Selected publications:
Rosen JN, Azevedo M, Soffar DB, Boyko VP, Brendel MB, Schulman VK, Baylies MK. 2019. The Drosophila Ninein homologue Bsg25D cooperates with Ensconsin in myonuclear positioning. J Cell Biol. 2019 Feb 4;218(2):524-540. doi: 10.1083/jcb.201808176. Epub 2019 Jan 9. PMID:30626718. Cover image
Manhart A, Windner S, Baylies M, Mogilner A. 2018. Mechanical positioning of multiple nuclei in muscle cells. PLoS Comput Biol. 2018 Jun 11;14(6):e1006208. doi: 10.1371/journal.pcbi.1006208. eCollection 2018 Jun.PMID:29889846
Schulman K, Folker ES, Rosen JN, Baylies MK. 2014. Syd/JIP3 and JNK signaling are required for myonuclear positioning and muscle function. PLoS Genet. 10(12):e1004880. doi: 10.1371/journal.pgen.1004880. eCollection 2014 Dec. PMID: 25522254
Sarcomerogenesis
Skeletal muscle cells or myofibers are required for locomotion. Each myofiber contains many linear myofibrils, which span the entire length of the cell. Each myofibril, in turn, is composed of a repeated array of sarcomeres, the fundamental contractile units of the muscle cell. The coordinated contraction of all sarcomeres along the myofibril shortens the entire myofiber and produces mechanical force. Sarcomeres are conserved in both structure and function amongst metazoans and have been studied for decades.
Current projects focus how sarcomeres are assembled, incorporated into myofibrils and are maintained during muscle development, growth, and homeostasis.
Selected publications:
Balakrishnan, M., Yu, SF., Chin,SM., Soffar, DB, Windner, SE., Goode, BL., Baylies MK 2019 Cofilin loss in Drosophila contributes to myopathy through defective sarcomerogenesis and aggregate formation during muscle growth. bioRxiv doi: https://doi.org/10.1101/825448. In Revision at Cell Reports.
Human muscle derived from human Progenitor stem cells (hPSCs)
The Drosophila model system has given an unparallel way to identify critical genes and mechanisms that drive muscle formation and differentiation. To translate our studies to human muscle, we have been using satellite cell derived muscle progenitors or IPSC-derived muscle progenitors to make 3D muscle “organoids”. These allow us to study some of the early events in making human muscle that are not possible to study in the embryonic or fetal stages.
Current projects focus on differentiation hPSCS into multinucleated myofibers, with resident satellite cells, co-cultures with human motorneurons and fibroblasts, investigation of myoblast fusion and myonuclear movement, and organelle positioning.